The group aims at investigating 3D atomic structures, assembly processes and molecular interactions of biological systems, including functional amyloids, bacterial appendages and membrane proteins. We develop and apply solid-state NMR to capture structural and dynamic details at the atomic scale. Our work is funded by the ERC, ANR, IdEx-Bordeaux and industrial contracts. More details on the ERC project Weakinteract here.

Functional amyloids
Amyloids are proteins that can undergo a conformational change from a soluble, monomeric to an insoluble, polymeric state, defined by the formation of aggregates ranging from oligomers to protofilaments and fibrils. Several amyloid proteins have been associated with the propagation of neurodegenerative diseases. Recently, numerous amyloid proteins have been identified in mammalians, fungi, bacteria or plants as crucial molecular determinants in the execution of native and beneficial biological functions, these proteins are named “functional amyloids” (in contrast to pathological amyloids). Well-known examples of functional amyloids include bacterial curli, hydrophobins or yeast prions. Recently, the formation of high-order fibrillar amyloid assemblies has been discovered in several signalling pathways controlling immunity-related cell fate. The precise role of amyloids in cell fate pathways and the structural mechanisms related to the templating and propagation of amyloid-based assemblies in signal transduction is still poorly understood. In collaboration with the group of Sven Saupe, we investigate the structure-function relationship of several functional amyloids involved in programmed cell death pathways.
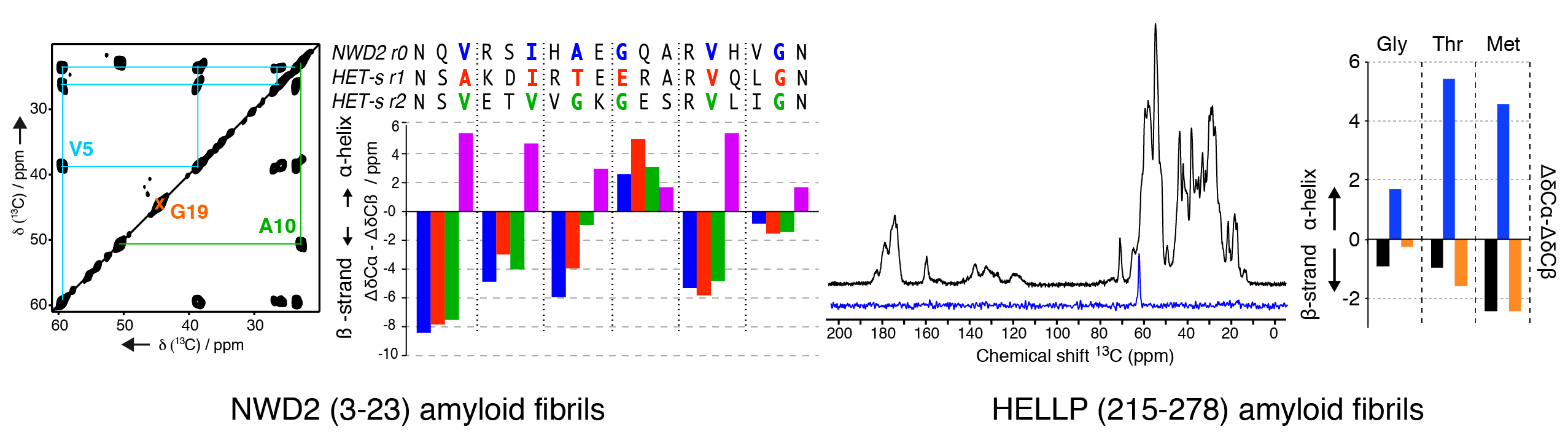
We have been investigating the structural architecture of the NWD2 protein, a NOD-like receptor with a N-terminal domain showing amyloid and prion properties. The NWD2 protein is encoded in a gene adjacent to het-S, a prion inducing programmed cell death in Podospora anserina. Using solid-state NMR, we determined the local conformation of the amyloid core based on strategic isotope labelling at key positions. Together with in vivo data from the Saupe group, it reveals that the NWD2 amyloid fibrils adopt a β-solenoid structure with a highly similar secondary structure compared to the HET-S/s-like systems. Involvement of amyloids in programmed cell death is not limited to fungal species, as recently uncovered for the RIP1/RIP3 necrosome in humans. We engaged into the structural characterization of the protein HELLP from C. globosum, sharing homology to the RHIM motif found in the execution of necroptotic cell death in humans. We recombinantly produced and purified HELLP (215-278) amyloid fibrils, and first solid-state NMR data indicate a rigid amyloid core in b-strand conformation.
References
Functional Amyloids in Health and Disease
Loquet A, Saupe SJ, Romero D.
J Mol Biol. 2018 Oct 12;430(20):3629-3630. doi: 10.1016/j.jmb.2018.07.024.
Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold
Daskalov A, Habenstein B, Martinez D, Debets AJ, Sabaté R, Loquet A, Saupe SJ.
PLoS Biol. 2015 Feb 11;13(2):e1002059. doi: 10.1371/journal.pbio.1002059
Daskalov A, Habenstein B, Sabaté R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ.
Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2720-5. doi: 10.1073/pnas.1522361113.
Solid-state NMR methods
We develop solid-state NMR methods to tackle complex biomolecular assemblies, to obtain atomic information on the structural architecture, molecular interactions and assembly mechanisms. We work on the development of new solid-state NMR experiments to improve sensitivity and spectral resolution, as well as on strategic isotope labeling schemes to improve the NMR analysis process. Ultra-fast magic-angle spinning NMR techniques are also investigated to improve sensitivity on complex biomolecular systems.
References
Tolchard J, Pandey MK, Berbon M, Noubhani A, Saupe SJ, Nishiyama Y, Habenstein B, Loquet A.
J Biomol NMR. 2018 Mar;70(3):177-185. doi: 10.1007/s10858-018-0168-3.
Bacterial Filamentous Appendages Investigated by Solid-State NMR Spectroscopy
Habenstein B, Loquet A.
Methods Mol Biol. 2017;1615:415-448. doi: 10.1007/978-1-4939-7033-9_29
Loquet A, Tolchard J, Berbon M, Martinez D, Habenstein B.
J Vis Exp. 2017
Atomic Structural Investigations of Self-Assembled Protein Complexes by Solid-State NMR
Loquet A, Tolchard J, Habenstein B.
Applications of NMR spectroscopy. in press
Stanek J, Andreas LB, Jaudzems K, Cala D, Lalli D, Bertarello A, Schubeis T, Akopjana I, Kotelovica S, Tars K, Pica A, Leone S, Picone D, Xu ZQ, Dixon NE, Martinez D, Berbon M, El Mammeri N, Noubhani A, Saupe S, Habenstein B, Loquet A, Pintacuda G.
Angew Chem Int Ed Engl. 2016 Dec 12;55(50):15504-15509. doi: 10.1002/anie.201607084
Solid-state NMR: An emerging technique in structural biology of self-assemblies
Habenstein B, Loquet A.
Biophys Chem. 2016 Mar;210:14-26. doi: 10.1016/j.bpc.2015.07.003.
Remorins and nanodomain interaction
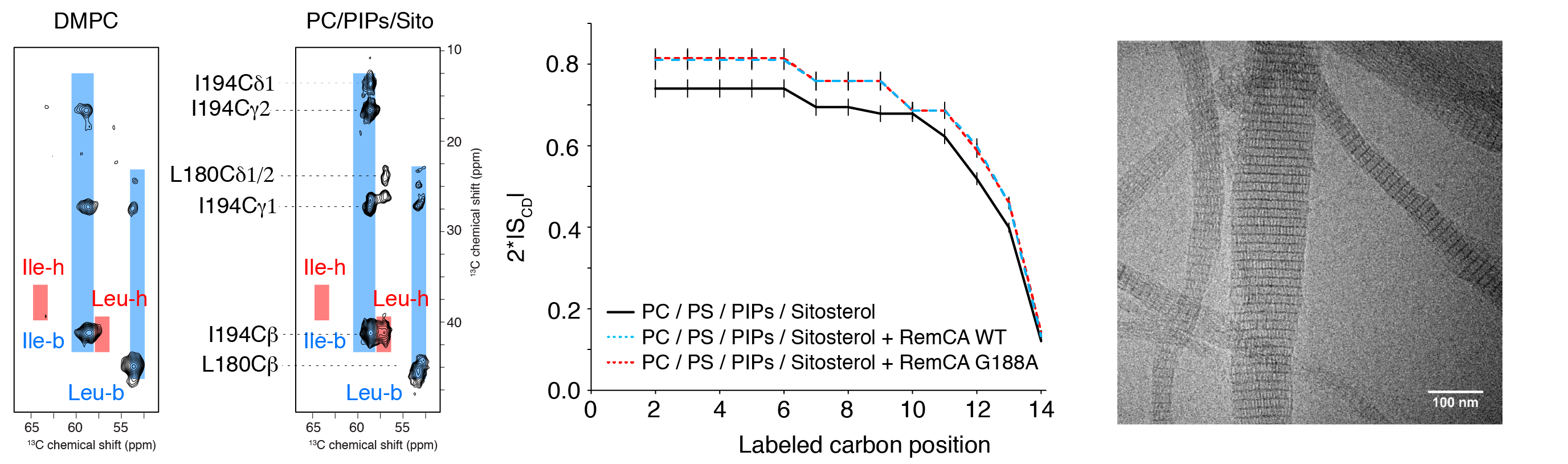
Membrane nanodomains represent an essential tool of the versatile membrane barriers to create platforms controlling cellular functions and interactions with the cellular environment. REMORINs are nanodomain-organized proteins located in the plasma membrane and involved in cellular responses in plants. In collaboration with S. Mongrand and co-workers (LBM CNRS) we have recently visualised by solid-state NMR (13C- and 2H-detected methods) that nanodomain localisation of REMORINs is mediated by sterols and PI4P and we could propose a model suggesting an unconventional binding mode via the C-terminal anchor domain. Using an ensemble of biophysical approaches, including solid-state NMR, cryo-EM and in vivo confocal imaging, we could also provide first insights into the role and the structural basis of REMORIN trimerization. REMORIN coiled-coil trimer formation regulates membrane recruitment and promotes REMORIN assembly in vitro into long filaments.
References
Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains.
Gronnier J, Crowet JM, Habenstein B, Nasir MN, Bayle V, Hosy E, Platre MP, Gouguet P, Raffaele S, Martinez D, Grelard A, Loquet A, Simon-Plas F, Gerbeau-Pissot P, Der C, Bayer EM, Jaillais Y, Deleu M, Germain V, Lins L, Mongrand S.
Elife. 2017 Jul 31;6. pii: e26404. doi: 10.7554/eLife.26404
Coiled-coil oligomerization controls nanodomain organization of the plasma membrane REMORINs.
Martinez D, Legrand A, Gronnier J, Decossas M, Gouget P, Lambert O, Berbon M, Verron L, Grélard A, Germain V, Loquet A, Mongrand S, Habenstein B
J Struct Biol. 2018 Feb 23. pii: S1047-8477(18)30046-7. doi: 10.1016/j.jsb.2018.02.003.
Lipid dynamics
The plasma membrane constitutes the interface between the cell and the external environment, playing a crucial role in response to external environmental factors. In order to insure essential processes such as signal transduction, membrane polarization or cellular traffic, membrane receptors adopt specific conformation, often lipid-dependent. Structural characterization of such membrane proteins is challenging to due to the nature of the membrane (insoluble, heterogeneous). We use solid-state NMR to understand conformational protein changes at the membrane interface, as well as to decipher lipid dynamics, in liposomes and in nanodiscs.
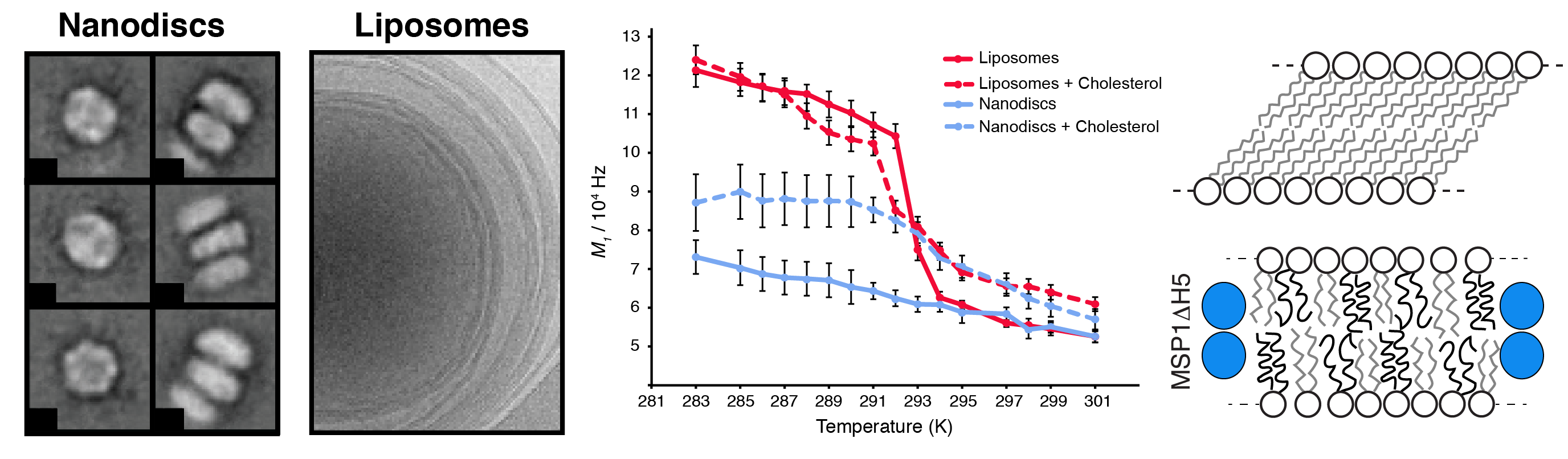
Nanodiscs represent a very promising technology, enabling the reconstitution of a soluble nano-object incorporated in a nanoscopic lipid bilayer. Tremendous advantages of nanodiscs (soluble, small, tunable, accessible to structural studies by high-resolution NMR and cryo-electron microscopy) make them one of the most promising tools to study membrane proteins at atomic resolution. The dynamic and structural behavior of the lipids incorporated in nanodiscs remains hardly investigated and poorly understood. For example, it remains unclear (i) if the membrane-like dynamics and thermotropism of lipids in nanodiscs are similar to the case of vesicles or cellular membranes. (ii) how the lipid-dynamics will adjust to the strictly confined membrane bilayer patch in nanodiscs upon transiting the gel-to-liquid phase transition temperature. (iii) how/if cholesterol changes the dynamics response of lipids when transiting the gel-to-liquid phase transition temperature. In collaboration with the group of Roland Riek (ETH Zurich) and Stefan Bibow (Biozentrum Basel), we have proposed a model of internal lipid dynamics inside nanodiscs and in comparison to liposomes.
References
Lipid Internal Dynamics Probed in Nanodiscs
Martinez D, Decossas M, Kowal J, Frey L, Stahlberg H, Dufourc EJ, Riek R, Habenstein B, Bibow S, Loquet A.
Chemphyschem. 2017 Jun 1. doi: 10.1002/cphc.201700450
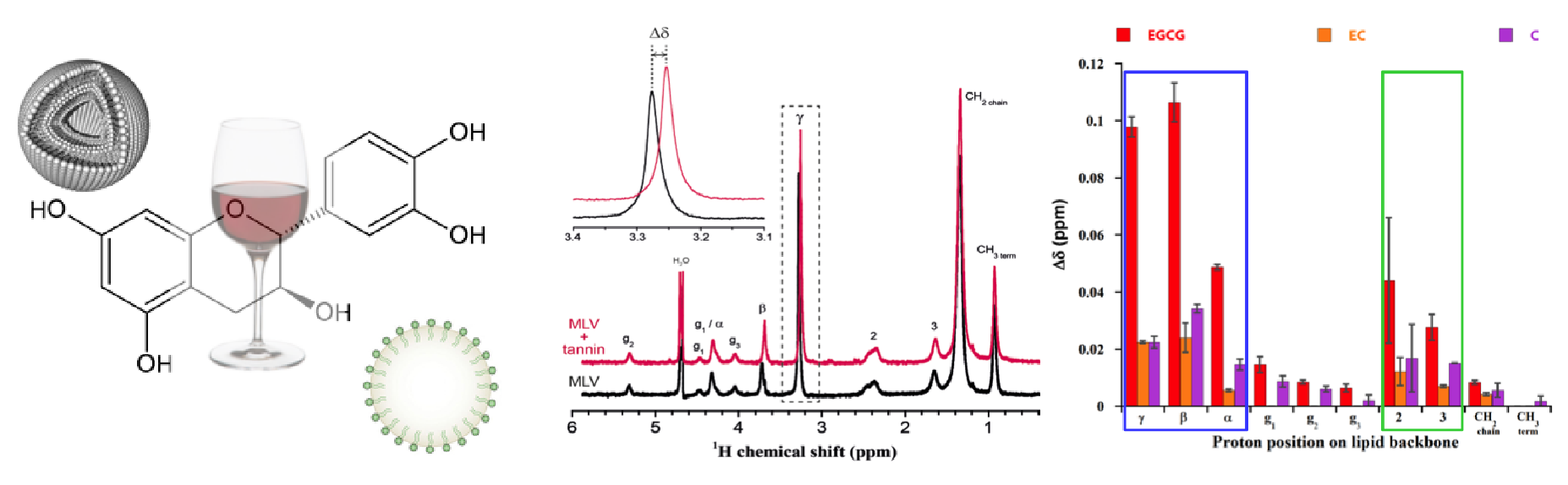
Tannin-lipid interactions
We study tannin-lipid interactions in the field of oenology and health. In oenology, tannins are responsible for the astringency and the bitterness of red wines. The former implicates an interaction between tannins and saliva proteins during mouth lubrication, while the latter results from an interaction between tannins and taste receptors. We investigate tannin-lipid interactions and the anti-oxidant efficiency of tannins in membranes by liquid and solid-state NMR.
References
Furlan AL, Saad A, Dufourc EJ, Géan J.
Biochimie. 2016 Nov;130:41-48. doi: 10.1016/j.biochi.2016.07.002
Furlan AL, Castets A, Nallet F, Pianet I, Grélard A, Dufourc EJ, Géan J.
Langmuir. 2014 May 20;30(19):5518-26. doi: 10.1021/la5005006
